Nicole Drakos
Research Blog
Welcome to my Research Blog.
This is mostly meant to document what I am working on for myself, and to communicate with my colleagues. It is likely filled with errors!
This project is maintained by ndrakos
MIRISim Troubleshooting Part II
In this post I’m going to troubleshoot why my MIRI simulations don’t have any visible sources. This is a continuation of Part I.
Test 2
For Test 2, I will expand upon the working Test 1 case (see the previous post), and include reading in APT and dither files
Code:
Part A
####################################################
# Check I can recover input galaxy from JWST pipeline
# conda activate mirisim
####################################################
import sys,os
sys.path.insert(0,'../../../Analysis')
from MyInfo import *
ra_offset = 150.11916667; dec_offset=2.20583333 #center, degrees
##########################################
#STEP A - Make fits file (Skip)
##########################################
##########################################
#STEP B - Visit info
##########################################
telescope_v3_pa = 293.09730273
num_dithers = 16 #for now this is hardcoded in. Assumes 16 entries in each visit.
# a) Read APT Pointing File
lookup = 'Observation'
visit_linenum = [] #save which line in the pointinf file each visit starts on
obs_num = [] # This is where I'll store the Observation number
#Read through file and save visit_linenum and obs_num
with open(pointing_file) as myFile:
for num, line in enumerate(myFile, 1):
if lookup in line:
visit_linenum.append(num)
obs_num.append(int(line.split()[-1]))
num_visits = len(visit_linenum)
#Save a list of dataframes for each observation
data_frames = []
for visit in visit_linenum:
df = pd.read_csv(pointing_file, skiprows = visit+1, nrows=num_dithers,delim_whitespace=True,index_col=False)
data_frames.append(df)
#Make a folder for each pointing
for obs in obs_num:
path = 'Obs' + str(obs)
if not os.path.exists(path):
os.makedirs(path)
#Save list of observations for this ATP File
myobsfile = pointing_file.split('/')[-1].split('.')[0]
np.savetxt(myobsfile+ '.obs', obs_num, fmt='%i')
##################################
# b) Make Dither Files
##################################
import pysiaf
from mirage.utils import siaf_interface
#Loop through visits
for i in range(num_visits):
obs = obs_num[i]
df = data_frames[i]
mypath = 'Obs' + str(obs)
telescope_v1_ra = df['RA'].iloc[0] # all RA must be the same in a dither sequence
telescope_v1_dec = df['Dec'].iloc[0] # all Dec must be the same in a dither sequence
#Only take Miri info and first exposure
df = df [(df['Aperture']=='MIRIM_ILLUM') & (df['Exp']==1)]
#Dithers
dithers = df[['DithX','DithY']].to_numpy() #arcsec
#convert to pixel coordinates
MIRI_SIAF = pysiaf.siaf.Siaf('MIRI')
MIRIM_FULL_SIAF = MIRI_SIAF.apertures['MIRIM_FULL']
local_roll, attitude_matrix, fullframesize, subarray_boundaries = \
siaf_interface.get_siaf_information(
MIRI_SIAF, 'MIRIM_FULL', telescope_v1_ra, telescope_v1_dec, telescope_v3_pa,
v2_arcsec = MIRIM_FULL_SIAF.V2Ref,
v3_arcsec = MIRIM_FULL_SIAF.V3Ref, # v2 v3 ref of MIRI FULL center
)
dithers_pix = MIRIM_FULL_SIAF.idl_to_sci(dithers[:,0],dithers[:,1])
X = dithers_pix[0] - MIRIM_FULL_SIAF.idl_to_sci(0,0)[0]
Y = dithers_pix[1] - MIRIM_FULL_SIAF.idl_to_sci(0,0)[1]
#Get information for wcs
col_ref, row_ref = 692.5, 511.5 #hardcoding this in right now
v2_ref, v3_ref = MIRIM_FULL_SIAF.sci_to_tel(col_ref+1, row_ref+1)
MIRIM_FULL_SIAF.set_attitude_matrix(attitude_matrix)
crval = MIRIM_FULL_SIAF.tel_to_sky(v2_ref, v3_ref)
crpix = MIRIM_FULL_SIAF.idl_to_sci(0.0, 0.0)
V3IdlYAngle= MIRIM_FULL_SIAF.V3IdlYAngle
VIdlParity = MIRIM_FULL_SIAF.VIdlParity
#save dither and pointing info
np.savetxt(mypath + '/dither.dat', np.array([X,Y]).T, fmt='%+.4f',delimiter=',')
np.savetxt(mypath + '/pointing.dat',np.array([telescope_v1_ra,telescope_v1_dec,telescope_v3_pa,local_roll,crval[0],crval[1],crpix[0],crpix[1],V3IdlYAngle,VIdlParity ]))
Part B
mport sys,os
sys.path.insert(0,'../../../Analysis')
from MyInfo import *
ra_offset = 150.11916667; dec_offset=2.20583333 #center, degrees
##########################################
#STEP C - Source and config files
##########################################
from mirisim.config_parser import SceneConfig, SimConfig
from mirisim.skysim import Background, Point, Galaxy, Skycube
from mirisim.skysim import sed
myobsfile = pointing_file.split('/')[-1].split('.')[0]
myobsfile = myobsfile+ '.obs'
obs_num = np.loadtxt(myobsfile)
for obs in obs_num:
mypath = 'Obs' + str(int(obs)) + '/'
# Source File - just put on 1 sersic galaxy in centre
bg = Background(level = 'low', gradient = 5., pa = 45.,centreFOV=(0,0))
#gal_loc = (3600*ra_offset,3600*dec_offset)
galaxy = Galaxy(Cen = (0,0),n=1.,re=5.,q=0.4,pa=0)
PL = sed.PLSed(alpha = 0, flux = 1e7, wref = 10.) #reference flux [microJy] at wavelength [microns]
galaxy.set_SED(PL)
targetlist = [galaxy]
scene_config = SceneConfig.makeScene(loglevel=0,background=bg,targets = targetlist)
scene_config.write(mypath+'scene.ini',overwrite=True)
# Config File
sim_config = SimConfig.makeSim(
name = "mirisim", # name given to simulation
scene = "scene.ini", # name of scene file to input
rel_obsdate = 0.0, # relative observation date (0 = launch, 1 = end of 5 yrs)
POP = 'IMA', # Component on which to center (Imager or MRS)
ConfigPath = 'IMA_FULL', # Configure the Optical path (MRS sub-band)
Dither = True, # Dither
StartInd = 1, # start index for dither pattern
DitherPat = 'dither.dat', # dither pattern to use
filter = 'F770W', # Imager Filter to use
ima_mode = 'FAST', # Imager read mode (default is FAST ~ 2.3 s)
ima_frames = 45, # number of groups (for MIRI, # Groups = # Frames)
ima_integrations = 2, # number of integrations
NDither = 4, # number of dither positions
ima_exposures = 8, # number of exposures
readDetect = 'FULL', # Portion of detector to read out
disperser = 'SHORT', # [NOT USED HERE]
detector = 'SW', # [NOT USED HERE]
mrs_mode = 'SLOW', # [NOT USED HERE]
mrs_exposures = 2, # [NOT USED HERE]
mrs_integrations = 3, # [NOT USED HERE]
mrs_frames = 5, # [NOT USED HERE]
)
sim_config.write(mypath+'config.ini',overwrite=True)
##########################################
#STEP D - Run MIRISim
##########################################
from mirisim import MiriSimulation
current_path = os.path.abspath(os.getcwd())
for obs in obs_num:
mypath = current_path + '/Obs' + str(int(obs)) + '/'
os.chdir(mypath)
mysim = MiriSimulation.from_configfiles('config.ini')
mysim.run()
Plots:
Here are the different dithers for the first visit:
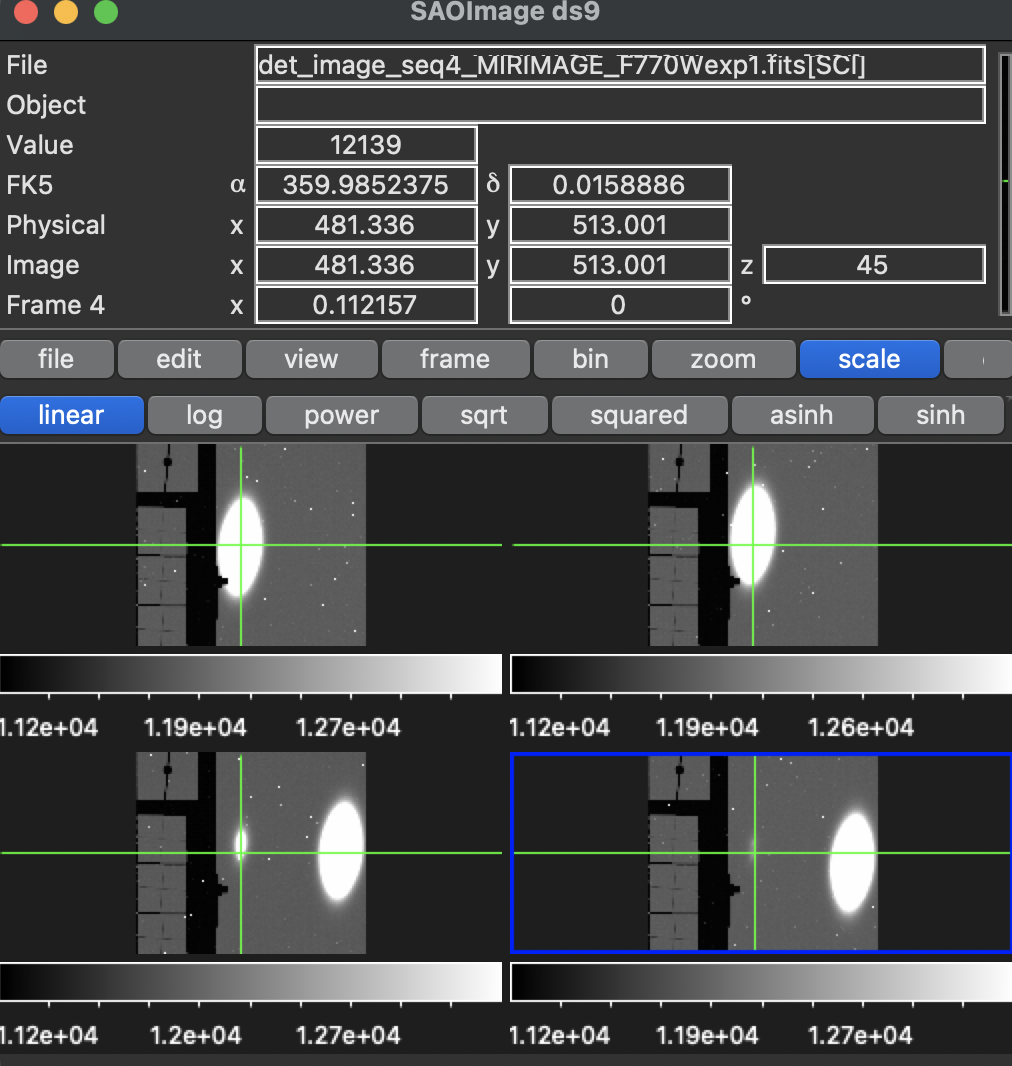
Note that the dither pattern given is (in pixels:)
-224.2712,-9.6723 -221.4668,+44.1934 +224.2700,+10.3781 +221.1110,-43.5871
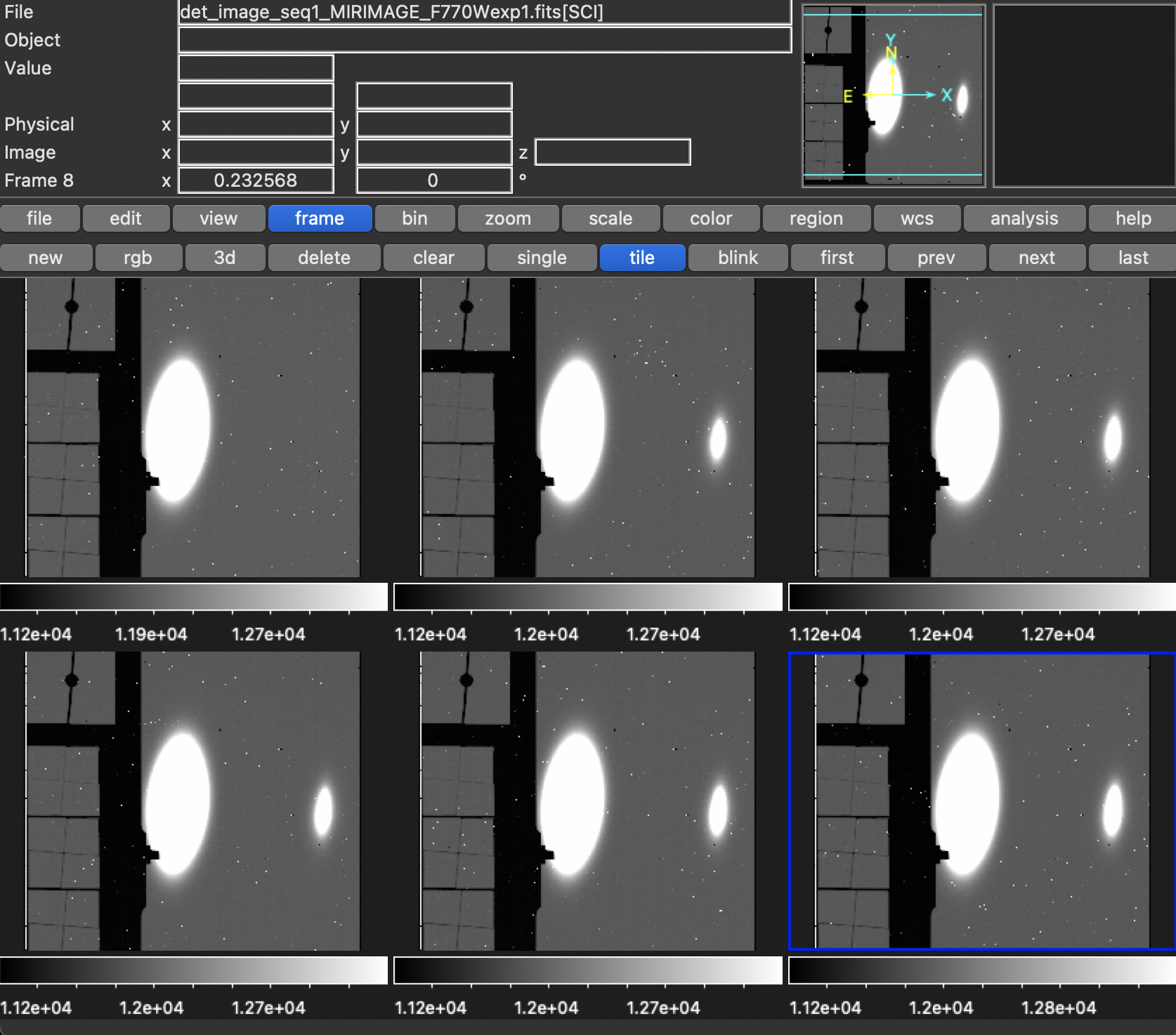
Here is the first image from each the 6 visits:
Notes:
- The dither patterns look right. As posted previously, the “centre” of the images is at (688.5,511.5) pixels, and the dither pattern was written in the previous section.
- The observations are all centred the same, which is how it should be since I gave them all the same scene. For the actual mock observations, I will centre the cutout of the scene based on the pointing.
- There is a bright spot on the raw images, that isn’t the one source put in. I’m not too worried about this though, since the image IS unrealistically bright. I believe we saw something similar in the NIRCam images, when we had a star that was too bright in them.
- I do need to check to make sure I am interpreting the pointing of the COSMOS APT files correctly, but at least now I am sure of how the input-> output is treated!
Next Steps
- Test 3: Save the cutouts of the scenes from the FITS files (i.e. include some of step A).
- Check: how do galaxy fluxes compare to DREaM?
- Make sure that Steps A-C are running correctly
- Test 4: Look into the “Run” step, make sure I am setting the WCS information correctly